Phylogeography: Evolution and climat
The genetic and demographic processes, especially speciation, that have shaped the geographic distribution and current structuring of populations, along with paleographic factors, are particularly valuable for predicting emerging viral pathogens.
The geographic distribution of species is of great interest to ecologists. In this project, we introduce a novel algorithm designed to analyze the information content of multiple sequence alignments in relation to geographic distribution. This algorithm employs a sliding window approach and utilizes the Robinson-Foulds distance to identify protein regions that are specific to carnivorous species, allowing them to adapt to their environment.
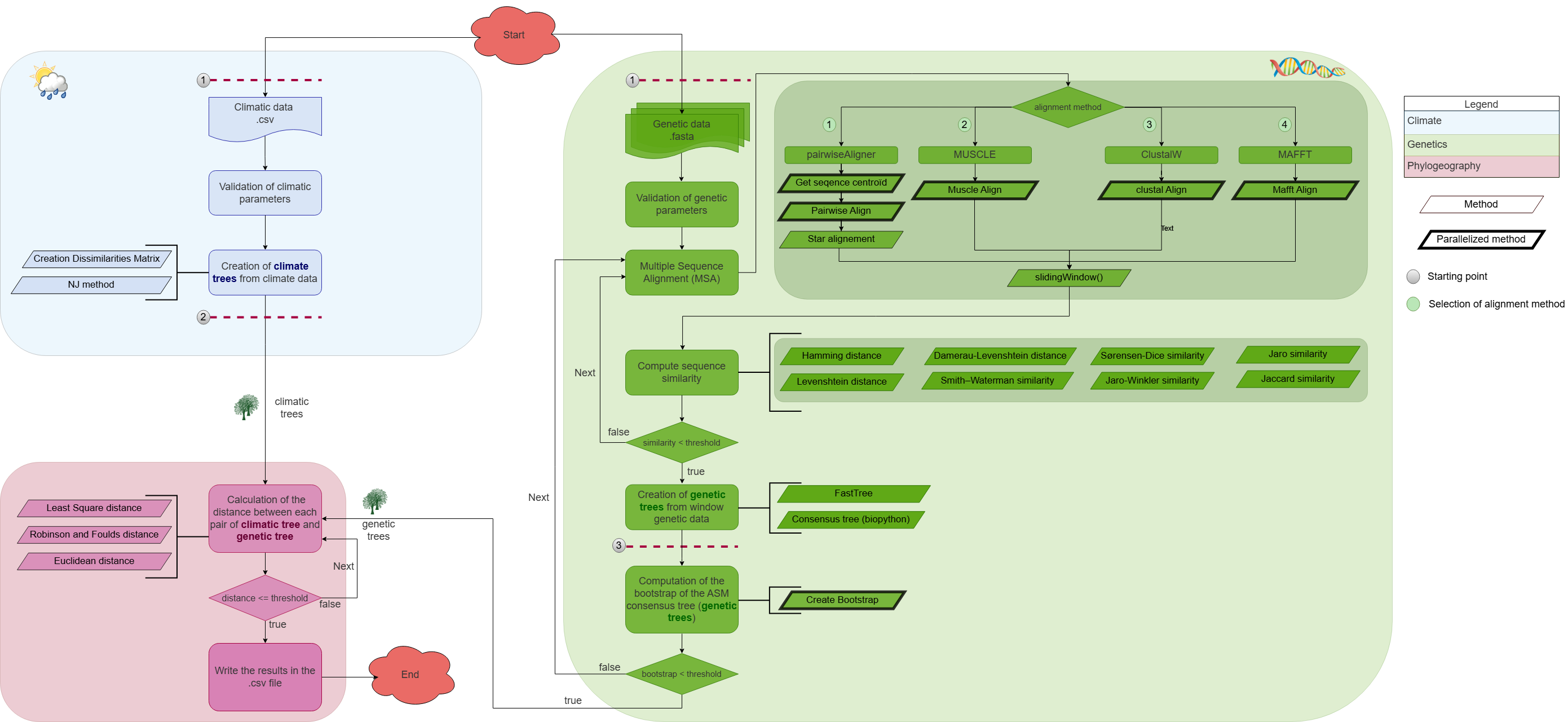
Source : aPhyloGeo on GitHub
Related publication:
- Chenoweth Galaz, A. L., & Tahiri, N. (2025). aPhyloGeo: a Python application for correlating genetic and climatic conditions. Bioinformatics, 41(11), btaf574.
- Li, W. & Tahiri, N. (2024). Host-Virus Cophylogeny Trajectories: Investigating Molecular Relationships between Coronaviruses and Bat Hosts. Viruses, 16(7), p.1133.
- Gagnon, J. & Tahiri, N. (2024). Ecological and Spatial Influences on the Genetics of Cumacea (Crustacea: Peracarida) in the Northern North Atlantic. roceeding in SciPy, pp.196-215, Tacoma, WA, USA
- Li, W. & Tahiri, N. (2023). aPhyloGeo-Covid: A Web Interface for Reproducible Phylogeographic Analysis of SARS-CoV-2 Variation using Neo4j and Snakemake. Proceeding in SciPy 2023, Auxtin, TX, USA
- Koshkarov, A., Li, W., Luu, M. L., & Tahiri, N. (2022). Phylogeography: Analysis of genetic and climatic data of SARS-CoV-2. Proceeding in SciPy 2022, Auxtin, TX, USA
- Tahiri, N., Badran, N., Dion-Phénix, H., Meniaï, I., & Makarenkov, V. (2017). Un nouvel algorithme pour l’analyse d’arbres phylogéographiques, Poster, 85e ACFAS.
FUNDING:
- Bourses de Stage de recherche Globalink
- Bourses du Fonds interculturel - Antje Bettin (Cycles supérieurs)
- Bourse d’excellence de l’UQAM pour cycles supérieurs (FARE)
- Bourse d’excellence Hydro-Québec et Fondation J. A. DeSève
- Bourse d’excellence de la fondation de l’UQAM (Prix Mario-Plouffe)
- Bourse d’excellence RBC Banque Royale